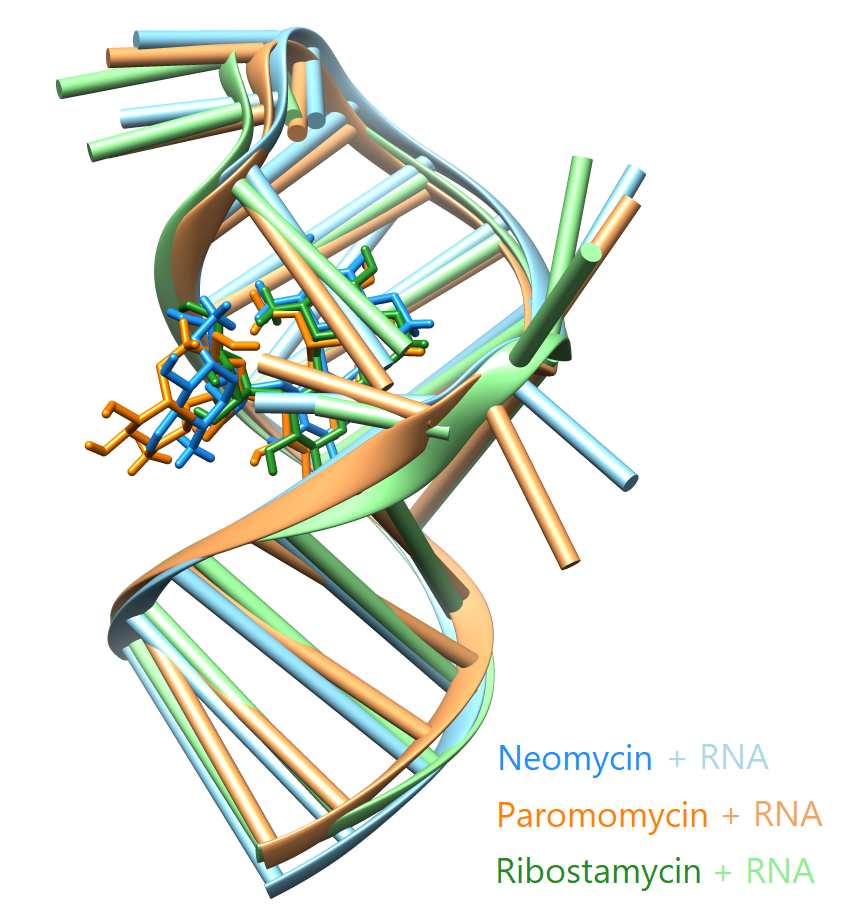
Molecular dynamics simulations of biological systems
Molecular dynamics simulations give an insight to the mechanisms of action of biological systems at atomic level of detail. An example of an intruiging system for that kind of study is an aminoglycoside riboswitch. Different ligands binding the N1 riboswitch, despite being very similar to one another, can affect the activity of the riboswitch in different ways. Neomycin and ribostamycin inhibit the expression of the genes located downstream from the riboswitch. Paromomycin binds the riboswitch but does not affect the gene expression. Here, we explain the molecular mechanisms that decide about the riboswitch activity in vivo.
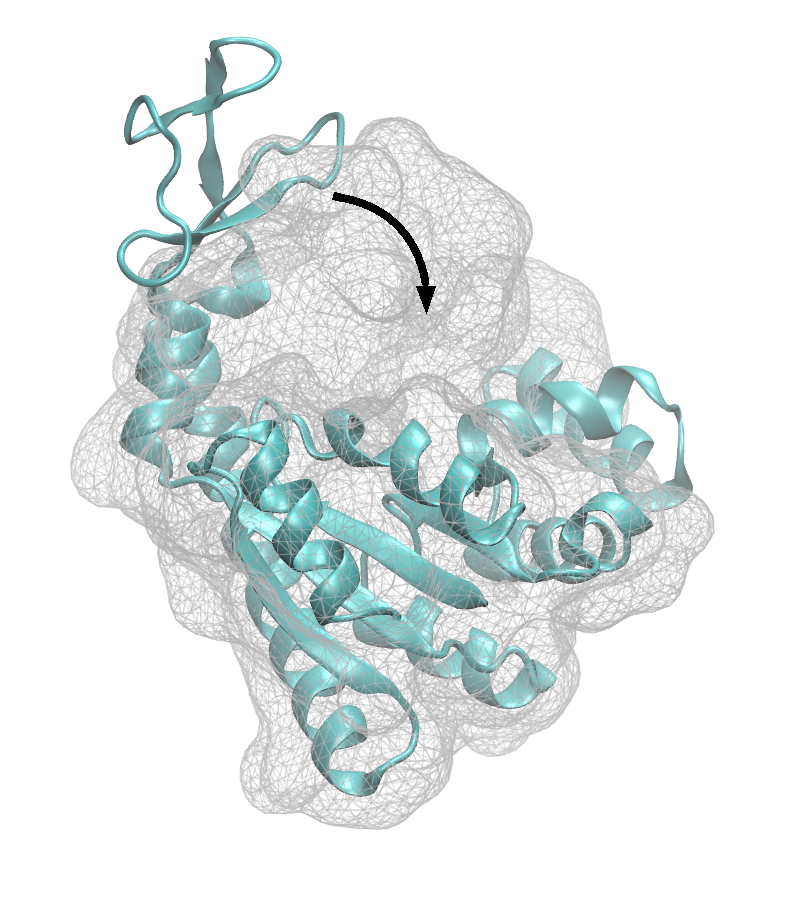
Molecular dynamics simulations with flexible fitting
Single-particle cryo-electron microscopy is a modern method in structural biology that provides many images of biological samples at almost sub-angstrom resolutions. Those images can be processed with 3D-reconstruction techniques to arrive at the 3D density maps of single molecules. Using homology modelling or molecule structures that are already available, we can fit an atomic structure to those density maps. However, the fit is usually not perfect. Molecular dynamics simulations with flexible fitting help to make this fit better.

Electron density and electrostatic potential maps of biomolecules
X-ray crystallography experiments can provide detailed view of the electron density of a molecule in a crystal. Similarly, single-particle cryo-electron microscopy reveals the electrostatic potential features of the studied molecule. Calculation of the detailed electron density and electrostatic potential maps of biomolecules, using advanced aspherical models, is able to provide even more detailed information about the functionally important molecular fragments.